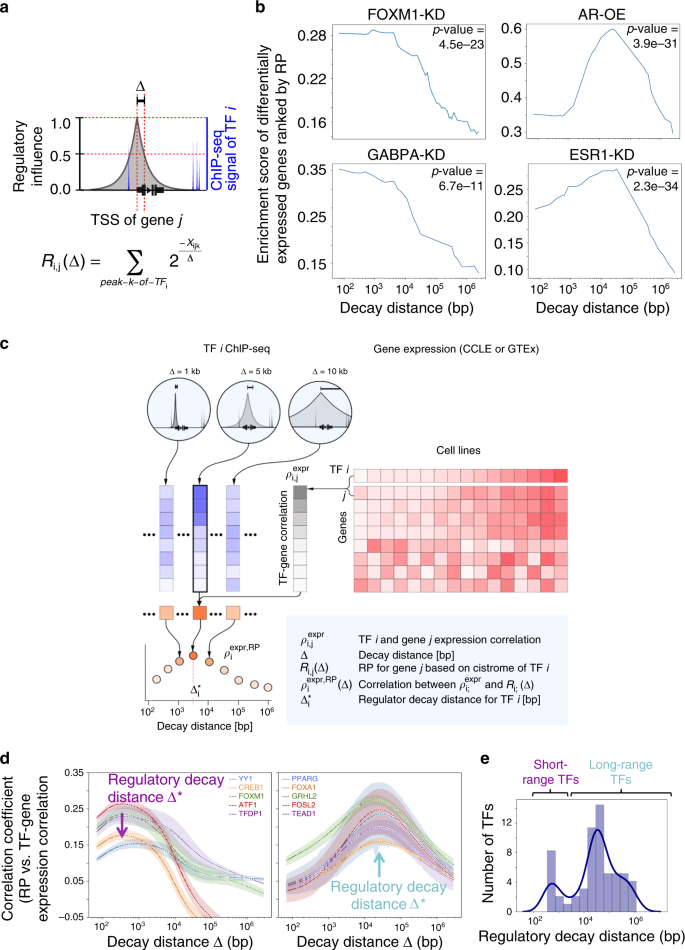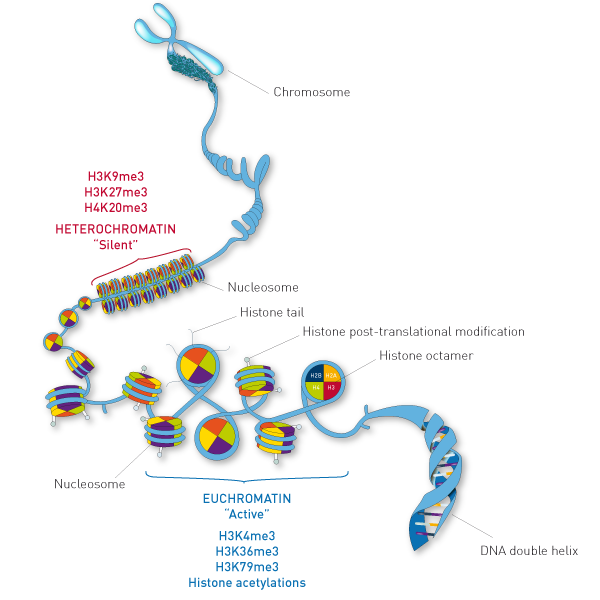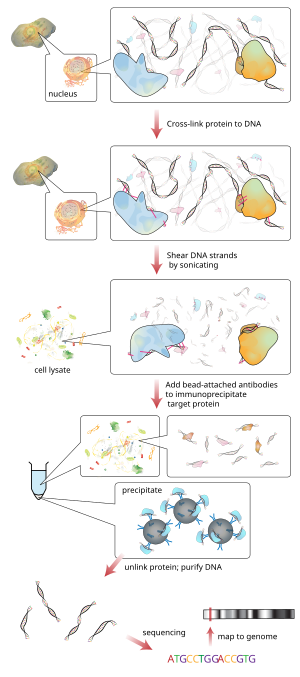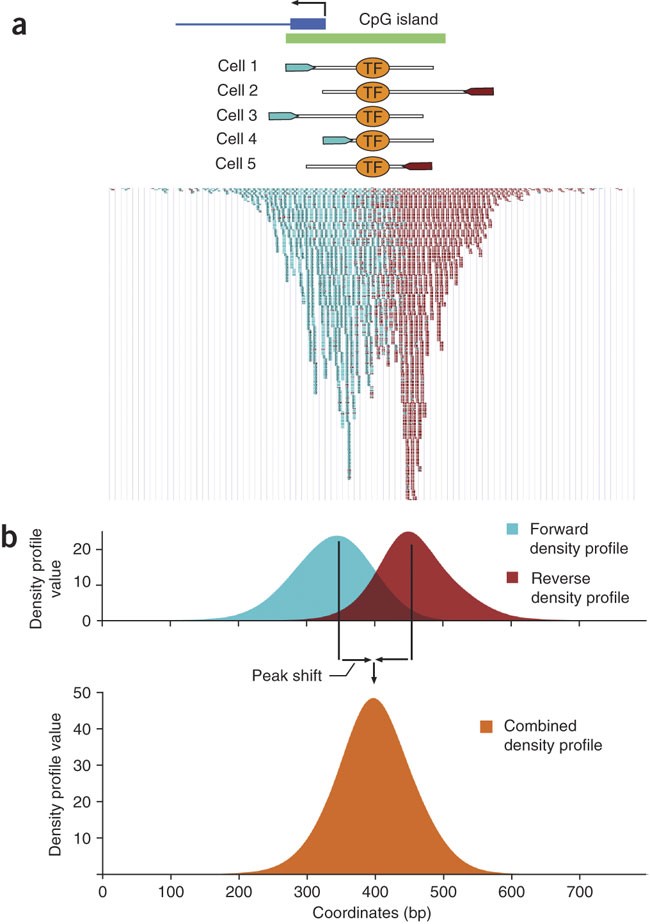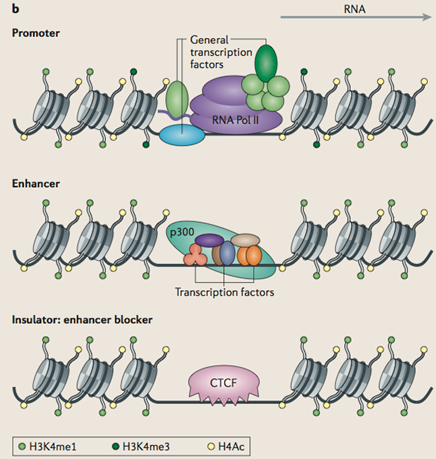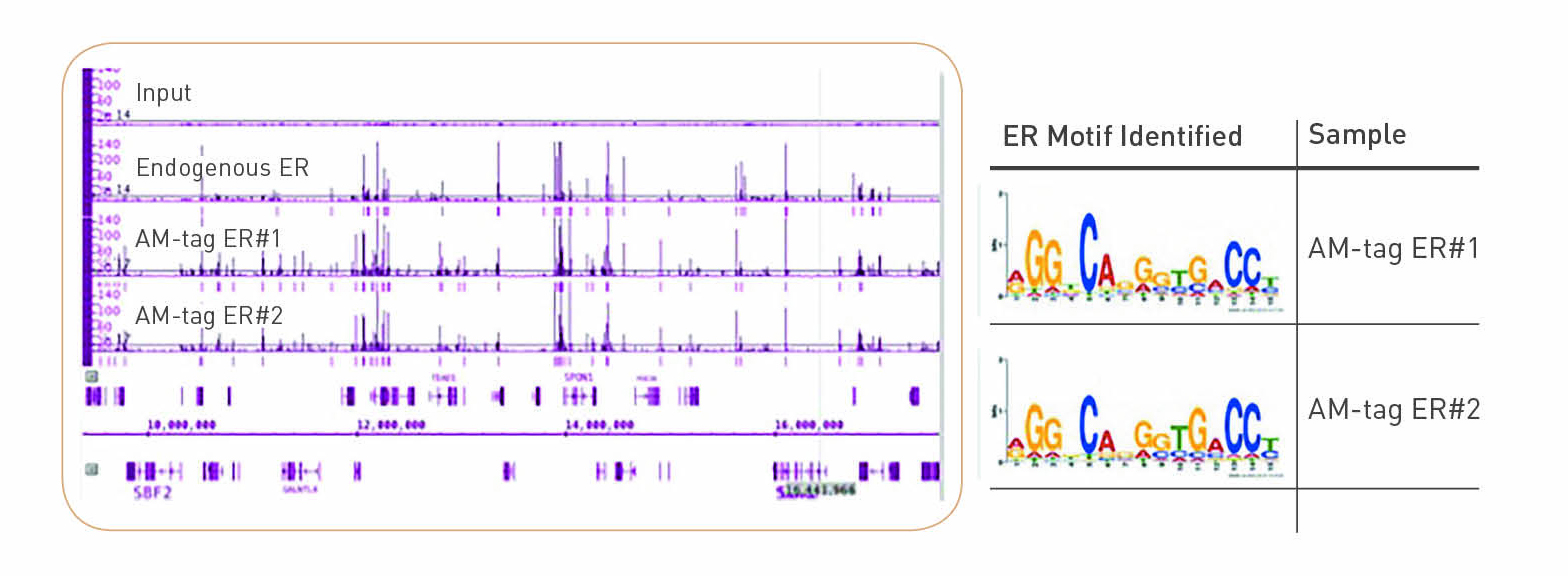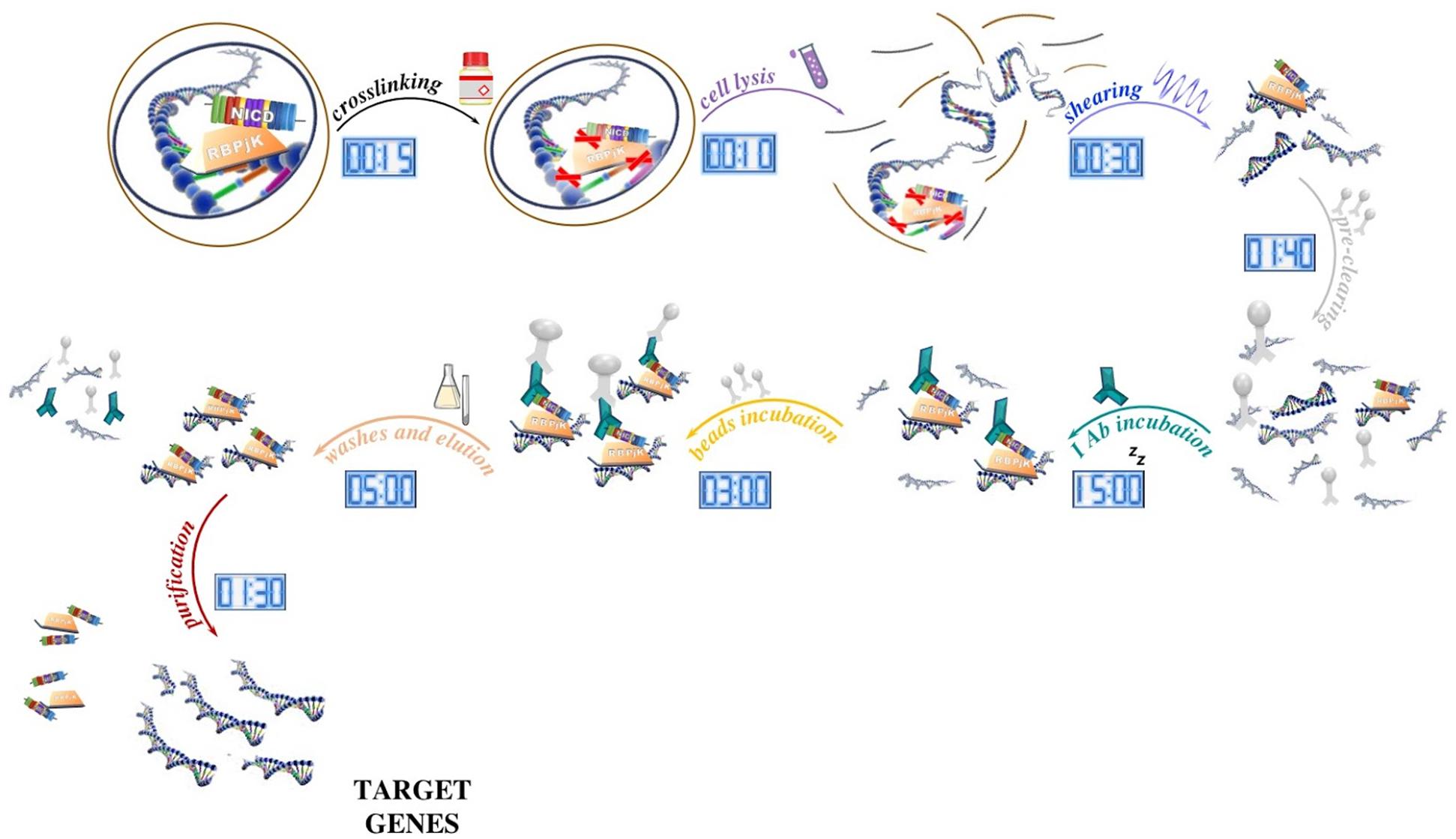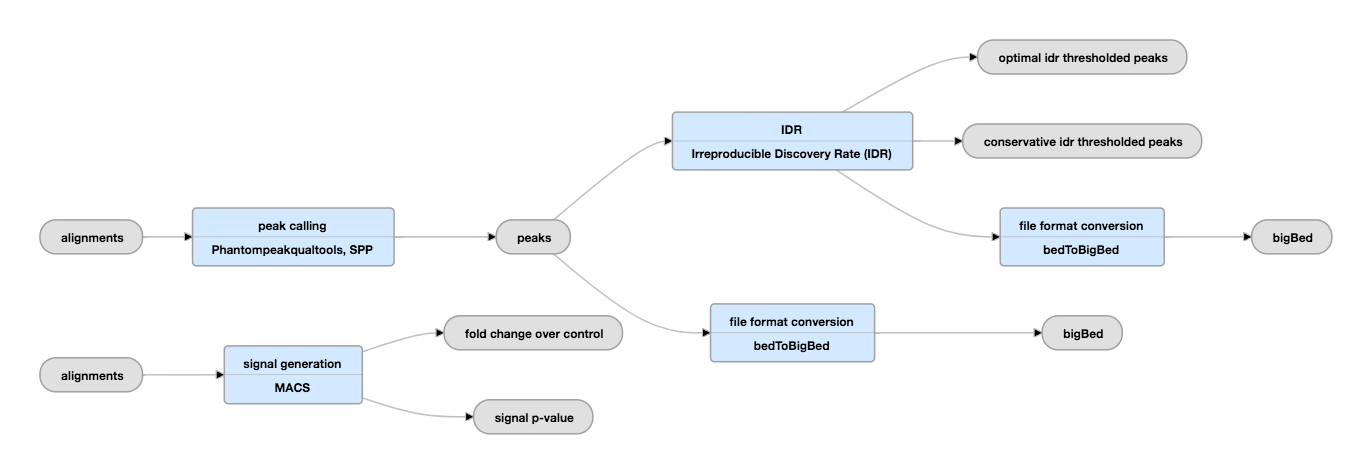
Optimized ChIP-seq method facilitates transcription factor profiling in human tumors | Life Science Alliance

Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells - ScienceDirect
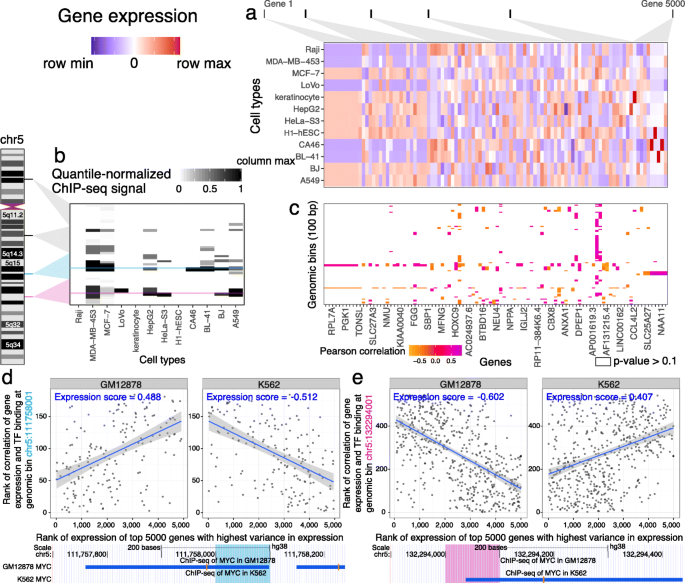
Virtual ChIP-seq: predicting transcription factor binding by learning from the transcriptome | Genome Biology | Full Text

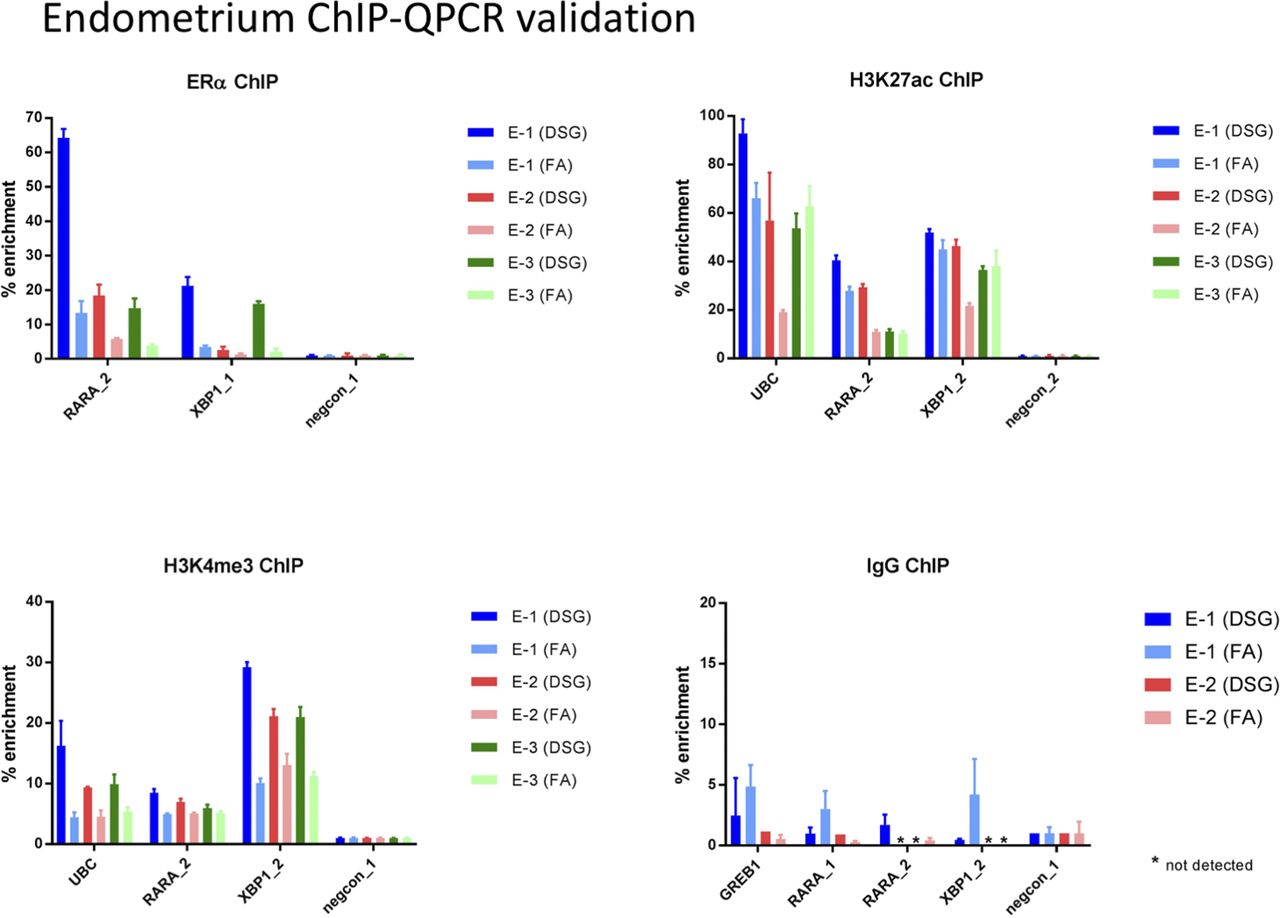
Validation of small-scale transcription factor ChIP-seq. (A) Profiles... | Download Scientific Diagram
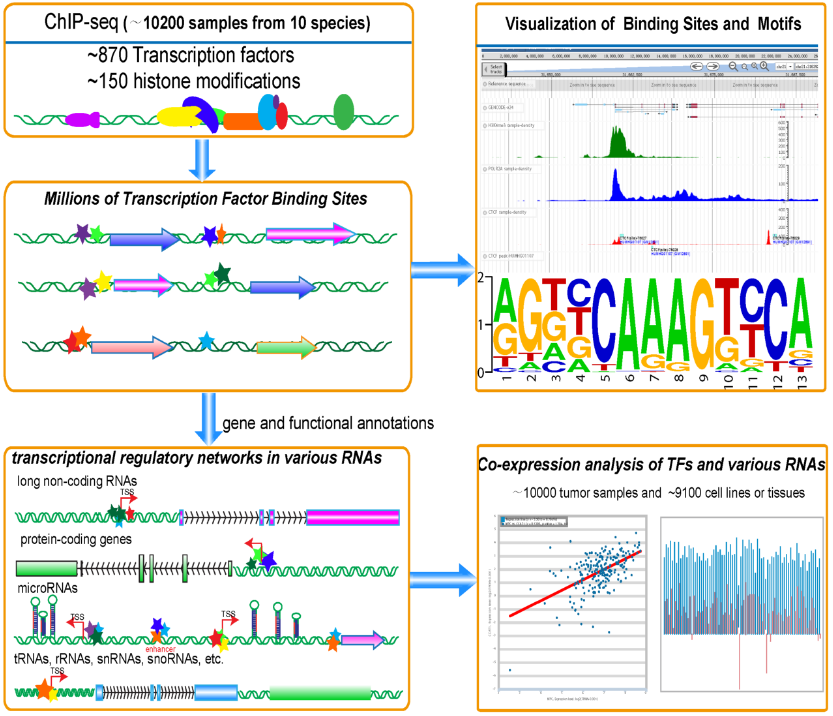
Transcription Factor Binding Sites, Motifs and Expression Profiles from ~10200 ChIP-seq and ~20000 RNA-seq samples

10,000 cell histone mark and transcription factor ChIP-seq. (A and B)... | Download Scientific Diagram

Chromatin Haiku on Twitter: "UV-crosslinking Transcription factor ChIP-seq New binding sites found #ChromatinHaiku #TxnJC https://t.co/9RuBoYUIyV https://t.co/FB4WLUKCMC" / Twitter

Optimized ChIP-seq method facilitates transcription factor profiling in human tumors | Life Science Alliance
![PDF] CisMapper: predicting regulatory interactions from transcription factor ChIP-seq data | Semantic Scholar PDF] CisMapper: predicting regulatory interactions from transcription factor ChIP-seq data | Semantic Scholar](https://ai2-s2-public.s3.amazonaws.com/figures/2017-08-08/9f500e90b2d7cedf7ced9593b62f9b2329fce216/3-Figure1-1.png)
PDF] CisMapper: predicting regulatory interactions from transcription factor ChIP-seq data | Semantic Scholar
The Role of Genome Accessibility in Transcription Factor Binding in Bacteria | PLOS Computational Biology
Detection of cooperatively bound transcription factor pairs using ChIP-seq peak intensities and expectation maximization | PLOS ONE

Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data

Validation of small-scale transcription factor ChIP-seq. (A) Profiles... | Download Scientific Diagram